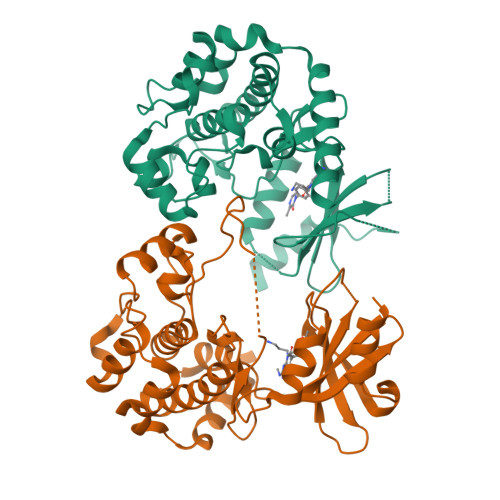
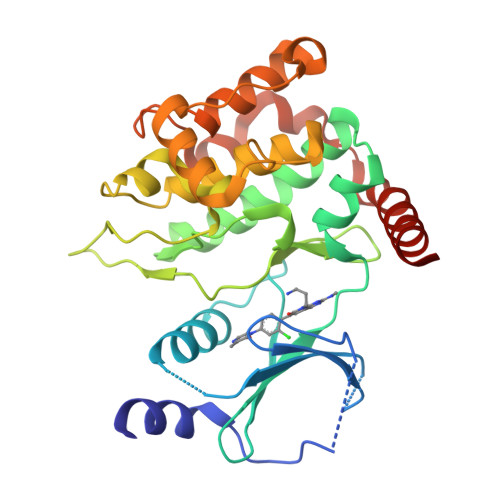
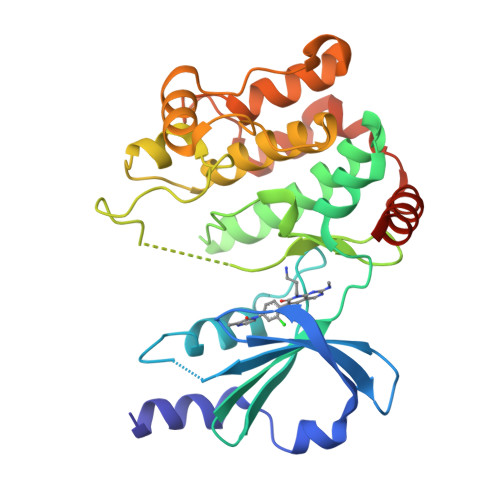
Chemically Diverse Group I p21-Activated Kinase (PAK) Inhibitors Impart Acute Cardiovascular Toxicity with a Narrow Therapeutic Window.
Rudolph, J., Murray, L.J., Ndubaku, C.O., O'Brien, T., Blackwood, E., Wang, W., Aliagas, I., Gazzard, L., Crawford, J.J., Drobnick, J., Lee, W., Zhao, X., Hoeflich, K.P., Favor, D.A., Dong, P., Zhang, H., Heise, C.E., Oh, A., Ong, C.C., La, H., Chakravarty, P., Chan, C., Jakubiak, D., Epler, J., Ramaswamy, S., Vega, R., Cain, G., Diaz, D., Zhong, Y.(2016) J Med Chem 59: 5520-5541
- PubMed: 27167326
- DOI: https://doi.org/10.1021/acs.jmedchem.6b00638
- Primary Citation of Related Structures:
5IME - PubMed Abstract:
p21-activated kinase 1 (PAK1) has an important role in transducing signals in several oncogenic pathways. The concept of inhibiting this kinase has garnered significant interest over the past decade, particularly for targeting cancers associated with PAK1 amplification. Animal studies with the selective group I PAK (pan-PAK1, 2, 3) inhibitor G-5555 from the pyrido[2,3-d]pyrimidin-7-one class uncovered acute toxicity with a narrow therapeutic window. To attempt mitigating the toxicity, we introduced significant structural changes, culminating in the discovery of the potent pyridone side chain analogue G-9791. Mouse tolerability studies with this compound, other members of this series, and compounds from two structurally distinct classes revealed persistent toxicity and a correlation of minimum toxic concentrations and PAK1/2 mediated cellular potencies. Broad screening of selected PAK inhibitors revealed PAK1, 2, and 3 as the only overlapping targets. Our data suggest acute cardiovascular toxicity resulting from the inhibition of PAK2, which may be enhanced by PAK1 inhibition, and cautions against continued pursuit of pan-group I PAK inhibitors in drug discovery.
Organizational Affiliation:
Shanghai Chempartner Inc. , 998 Halei Road, Zhangjiang Hi-Tech Park, Pudong New Area, Shanghai 201203, People's Republic of China.